Huidong Chen
Machine Learning Enthusiast
About Me
A passionate machine learning enthusiast constantly exploring the cutting-edge advancements in artificial intelligence, deep learning models, and their real-world applications.
- Artificial Intelligence
- Machine Learning
- Deep Learning
- Generative AI
- Neural Networks
- GNN, LLM, CNN
-
PhD in Computer Science, 2012~2018
Tongji University, Shanghai, China
-
Visiting PhD student in Computational Biology, 2016~2017
Massachusetts General Hospital & Harvard Medical School, Boston, USA
-
Joint-training PhD student in Biostatistics and Computational Biology, 2015~2017
Dana-Farber Cancer Institute & Harvard T.H. Chan School of Public Health, Boston, USA
-
BSc in Computer Science, 2008~2012
Tongji University, Shanghai, China
Skills
Experience
Software
Featured Publications
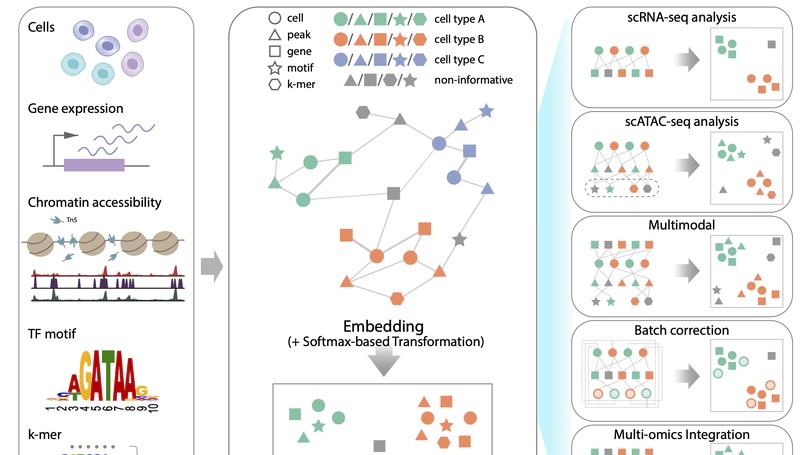
SIMBA is a method to embed cells along with their defining features such as gene expression, transcription factor binding sequences and chromatin accessibility peaks into the same latent space. The joint embedding of cells and features allows SIMBA to perform various types of single cell tasks, including but not limited to single-modal analysis (e.g. scRNA-seq and scATAC-seq analysis), multimodal analysis, batch correction, and multi-omic integration.